Welcome
We have a new group website available. Please check this website for more updated information of Jing Li lab:

We are a computational chemistry/biology research group in the Department of BioMolecular Sciences at the University of Mississippi School of Pharmacy. We are a computational chemistry/biology research group, and our research interests are largely directed towards understanding sequence-structure-dynamics-function relationship of biomolecules, the molecular mechanism of the dysfunctions of these biomolecules leading to diseases, and computer-aided drug discovery to treat these diseases. Inspired by the great physiological/biomedical significance of several families of membrane proteins, such as voltage-gated sodium (Nav) channels and g-amminobutyric acid type A (GABAa) receptors, we are particularly interested in the molecular mechanisms of disorders induced by missense mutations of these critical drug targets.

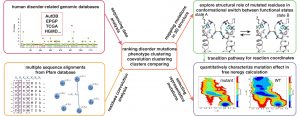
To open an intelligent and efficient route to illuminate the biological mechanisms of diseases in the era of big data, we use bioinformatics tools to extract disease-associated missense mutations and residue coevolution information from genomic sequencing data, and employ molecular dynamics simulation to study structural effects of diseases-causing mutations. Our overarching goal is to develop a computational framework as an “express” way connecting sequencing data analysis, structural biology, computational biophysics, and machine learning. It will significantly facilitate mechanism study guided by updated clinical data, and the translation of basic research into clinical applications.